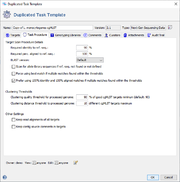
Target Scan Procedure Details
When WGS data is imported, SeqSphere+ scans the data for the target ref.-seqs. from the Task Template (using the integrated BLAST). If the scan finds a unique hit for a target, that reaches the thresholds that are specified here, this hit is imported as Target of the Task Entry.
The following parameters can be specified:
- Required identity to ref.-seq. (default for cgMLST: 90%)
- Defines the required percental identity of the BLAST hit compared to the ref.-seq. of this target.
- Required perc. aligned to ref.-seq. (default for cgMLST: 100%)
- Defines the required percental aligned overlap compared to the full length of the ref.-seq. of this target.
- Scan for allele library sequences if ref.-seq. not found or not defined (default for cgMLST: disabled)
- This option can be enabled to scan not only for the ref.-seq. of a target, but for all allele that already exist in the allele library for this target.
- Force using best match if multiple matches found within the thresholds (default for cgMLST: disabled)
- By default, only unique hits are imported. If multiple different hits were found that all reach the defined thresholds, none is imported. If this option is enabled always the best match (by score) is imported.
![]() Hint: For public Task Templates that were downloaded from the Task Template Sphere the thresholds are not editable.
Hint: For public Task Templates that were downloaded from the Task Template Sphere the thresholds are not editable.
Clustering Thresholds
Clustering thresholds are only available for cgMLST Task Templates. They are used by SeqSphere+ as default for MST Clusters, Early Warning Alerts, Local Single Linkage Clustering IDs, and for the Similar Samples Search.
![]() Hint: For public cgMLST Task Templates that were downloaded from the Task Template Sphere the thresholds are not editable. The quality threshold also defines if a sequence can be submitted to the nomenclature server. The clustering distance threshold also defines the cgMLST Complex Type (CT) distance.
Hint: For public cgMLST Task Templates that were downloaded from the Task Template Sphere the thresholds are not editable. The quality threshold also defines if a sequence can be submitted to the nomenclature server. The clustering distance threshold also defines the cgMLST Complex Type (CT) distance.
Two thresholds can be defined for clustering:
- Clustering quality threshold (default for cgMLST: 90)
- The quality threshold of percental good cgMLST targets that is required before the clustering is performed.
- Clustering distance threshold (default for cgMLST: empty)
- The distance threshold of maximum absolute different cgMLST targets, that is used to indicates epidemiological relationship between two Samples.
Other Settings
- Keep read alignments of all targets (default for cgMLST: disabled)
- By default, the read alignments for targets that pass the target QC procedure without errors or warnings are not kept. If this option is enabled, the read alignments are always kept.
- Keep contig source comments in targets (default for cgMLST: disabled)
- Can be enabled to store the original contig names where the target sequences were found during target scan procedure.