Please Note: Raven requires the ONT Data Assembly Module
Overview
Raven (citation) is a de novo assembler for long read data (e.g., Oxford Nanopore). It can be used in the assembling pipeline if the SeqSphere+ client is running on Linux or if the Windows Subsystem for Linux is installed on Windows. It is only available with the ONT Data Assembly Module.
See the page assembler evaluation for a detailed comparison of the performance.
Raven Assembling Options
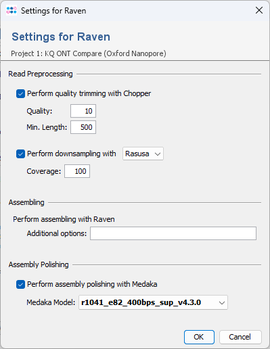
The assembler options dialog in the pipeline script allows to specify the pre- and postprocessing, and to set additional Raven parameters.
- Read trimming with Chopper (citation)
- Read downsampling with Rasusa (citation) or Filtlong (citation)
- Assembly polishing with Medaka (citation)
By default, the reads are trimmed with Chopper (quality 10, min length 500) and downsampled with Rasusa to 100x estimated coverage.
After assembling, medaka is used by default to polish the assembly. When confirming the project settings of the pipeline script, the assembler options dialog opens up and the newest available medaka model for dorado super accuracy basecalling (SUP) is preselected. Press OK to confirm these settings.