Please Note: Working with Plasmid Tables requires the Long-read Data Plasmid Transmission Analysis Module
The plasmid table displays plasmids from Mash Plasmid Typing searches, from Plasmid Early Warning Alerts, or from a snapshot of another plasmid table.
For each plasmid, there is a row that displays plasmid name, Sample ID, Project, Sample fields and mob-suite results.
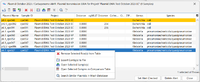
The following functions can be used:
-
 Export Table: Export the table.
Export Table: Export the table.
-
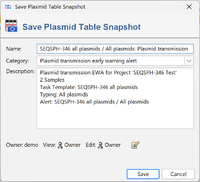
 Save Plasmid Table Snapshot: Create a snapshot of the plasmid table. The snapshots can be opened using the menu command Tools > Comparison Table. Note that it is not possible to search for similar plasmids when opening the snapshots.
Save Plasmid Table Snapshot: Create a snapshot of the plasmid table. The snapshots can be opened using the menu command Tools > Comparison Table. Note that it is not possible to search for similar plasmids when opening the snapshots.
-
 Add Additional Database Fields as Columns.
Add Additional Database Fields as Columns.
-
 Remove Selected Row(s) from table. Note: This will not remove the plasmids permanently from the database, only the rows from the table will be removed.
Remove Selected Row(s) from table. Note: This will not remove the plasmids permanently from the database, only the rows from the table will be removed.
-
 Remove Unselected Row(s) from Table. Note: This will not remove the plasmids permanently from the database, only the rows from the table will be removed.
Remove Unselected Row(s) from Table. Note: This will not remove the plasmids permanently from the database, only the rows from the table will be removed.
-
 Export Contigs to File: Reads the plasmid contigs from the Sample attachment and saves them to a FASTA-file.
Export Contigs to File: Reads the plasmid contigs from the Sample attachment and saves them to a FASTA-file.
-
 Open Selected Sample(s).
Open Selected Sample(s).
-
 Open Selected Sample(s) in Comparison Table.
Open Selected Sample(s) in Comparison Table.
-
 Visualize plasmids with pyGenomeViz. Metadata from the "Chromosome & Plasmid Overview" (rep(s) in green), "NCBI AMRFinderPlus" (AMR priority genes: red, other orange) and "CGE MobileElementFinder" (light blue) are included as genes in the visualization.
Visualize plasmids with pyGenomeViz. Metadata from the "Chromosome & Plasmid Overview" (rep(s) in green), "NCBI AMRFinderPlus" (AMR priority genes: red, other orange) and "CGE MobileElementFinder" (light blue) are included as genes in the visualization.
-
 Search Similar Plasmids in Mash Database.
Search Similar Plasmids in Mash Database.
Plasmid Tables that are created for similarity search or Plasmid Early Warning Alerts have the option to do a ![]() clustering of the plasmids in the table.
clustering of the plasmids in the table.